本期解读
题目:Integrative omic and transgenic analyses revealthe positive effect of ultraviolet-B irradiation on salvianolic acidbiosynthesis through upregulation of SmNAC1
期刊:The Plant Journal
影响因子:6.141/Q1
合作技术:iTRAQ定量蛋白组学
一、研究背景
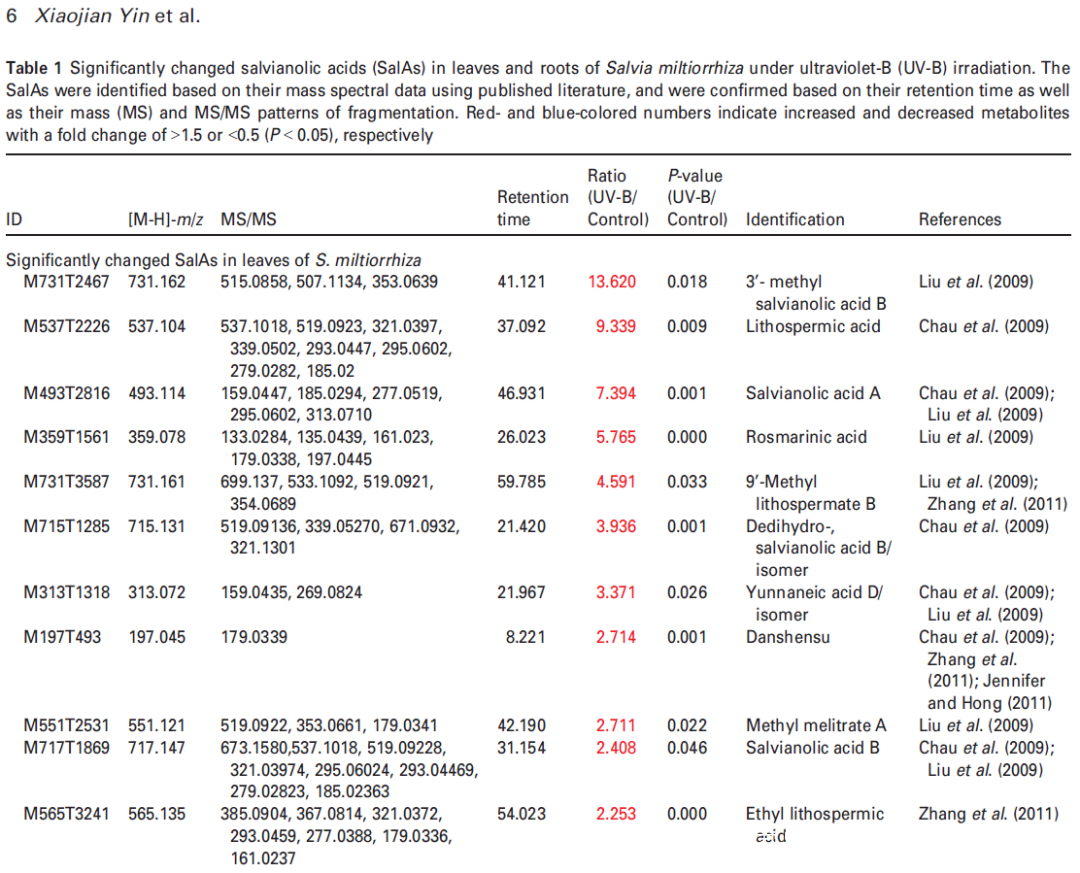
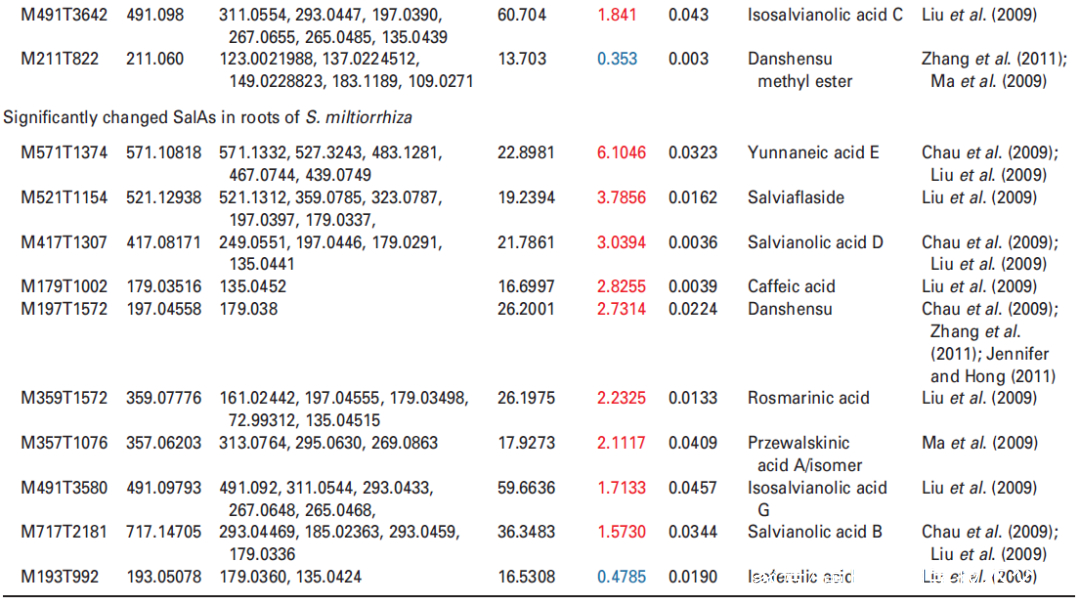
丹酚酸SalAs是丹参中重要的水溶性成分,有助于治疗脑血管和心血管疾病。据报道,在12种增加的丹酚酸中,丹酚酸B、丹酚酸A、迷迭香酸和丹参素对治疗脑血管疾病具有活性。最近,越来越多的研究旨在提高丹参中SalA的含量。并且已有研究表明,通过适当剂量的UV-B照射可以在体内增加SalA含量;然而,这种现象背后的机制仍然未知。
二、技术路线图
三、实验结果
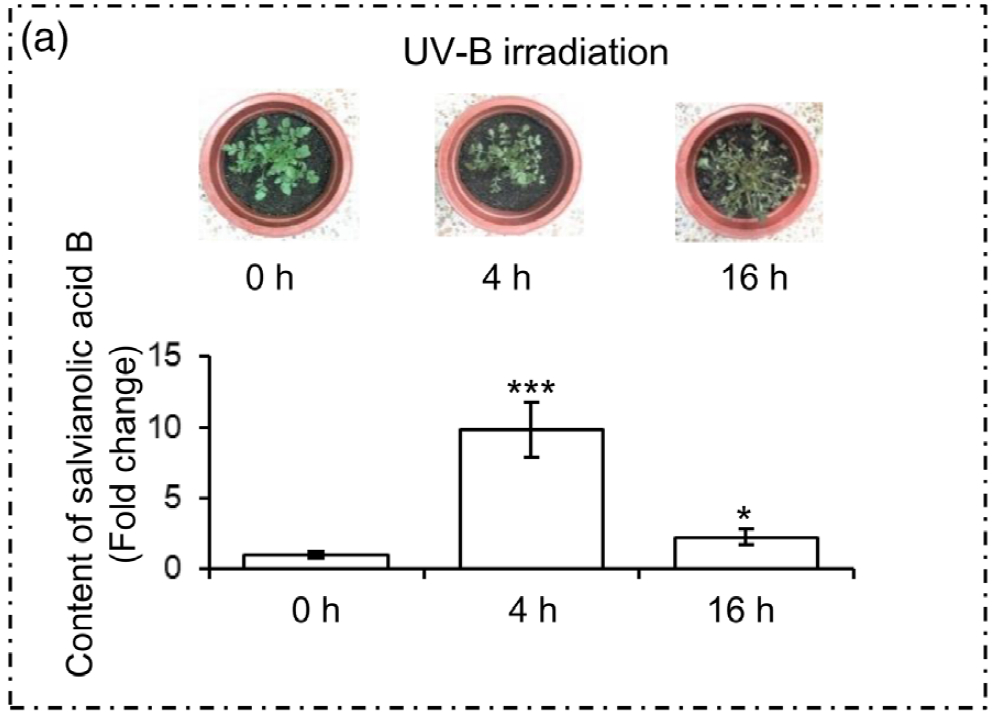
1. UV-B照射诱导丹参SalA合成的适宜时间选择
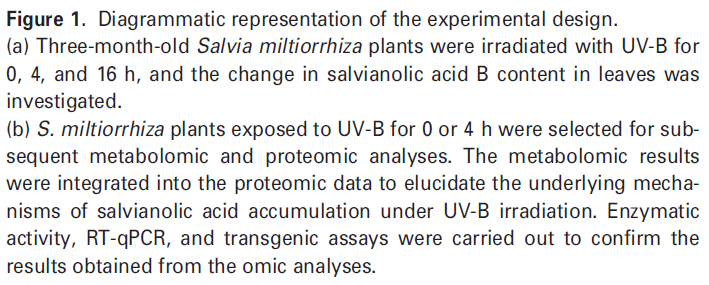
为了选择合适的 UV-B 照射条件,分析对比了在中度时长(4 小时)和长时间(16 小时)的UV-B照射后丹参的表型和丹酚酸的含量。结果显示,丹参在UV-B照射4小时后正常生长,但照射16小时后几乎死亡(图1(a)。与对照组相比,UV-B 照射4小时后丹酚酸水平显著增加,处理16小时后仅略有增加(图1(a)。因此,选择暴露于UV-B照射0小时(对照)和4小时(UV-B处理)的丹参用于响应UV-B照射的进一步生理、生化和分子研究(图 1(b))。
2. UV-B辐射下丹参中SalAs含量变化的代谢组学检测
研究者利用比较代谢组学方法系统地分析了0小时(对照)和4小时UV-B照射的丹参叶和根中SalAs的含量(图1(b))。代谢组学结果表明,经UV-B照射的叶子或根部获得的总离子色谱图与未照射样品的总离子色谱图明显不同(图 S1(a,b))。从丹参的叶和根中分别鉴定出1255和1853个离子(图2(a)),并且通过主成分分析(PCA)将UV-B照射的样品与对照样品清楚地聚集在一起(图S2(a,b))。使用Fold-Change ≥1.5(显著增加;P≤0.05)或≤0.5(显著减少;P≤0.05)的筛选标准,245和325个离子(共570个)分别在叶片中增加和减少,90和91个离子(共181个)分别在根中增加和减少(图2(a))。值得注意的是,已被证实在SalA生物合成中发挥重要作用的茉莉酸(JA)和SA的含量在丹参叶片中也确实显著增加(图S5和6;表S3)。
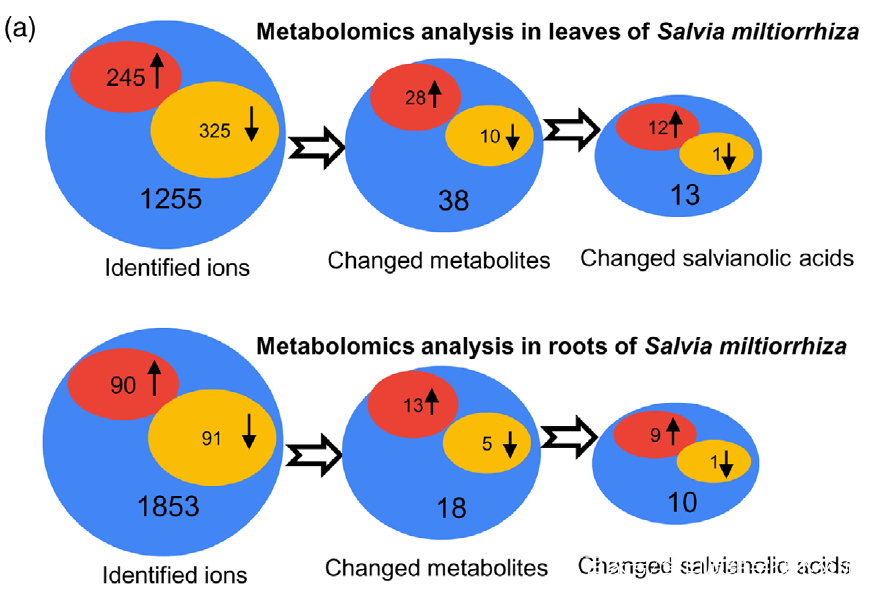
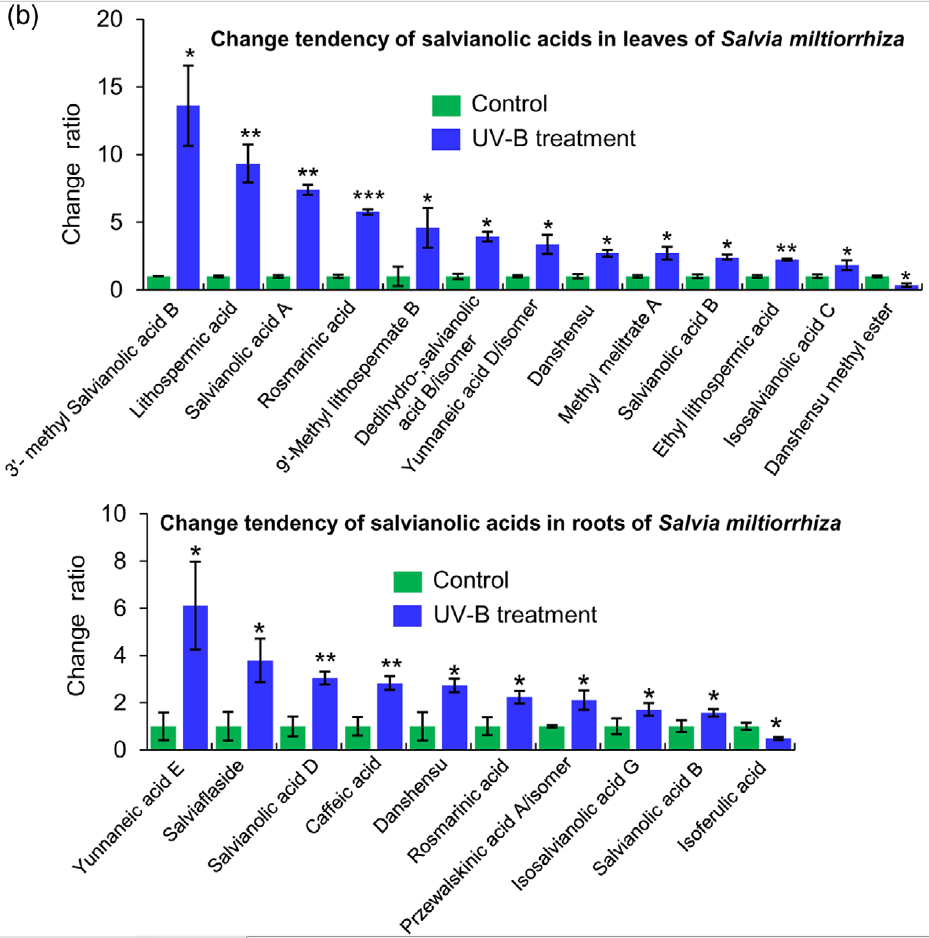
Figure 2. Effects of UV-B exposure on salvianolic acid (SalA) contents in leaves and roots of Salvia miltiorrhiza. Three-month-old S. miltiorrhiza plants were either untreated (controls) or treated with UVB exposure for 4 h. Metabolites extracted from leaves or roots were subjected to metabolomic analysis using untargeted ultra-performance liquid chromatography-quadrupole-time of flight mass spectrometry (UPLC-Q/TOF-MS).
(a) The statistical numbers of identified ions, metabolites, and SalAs from leaves and roots are listed. The arrows facing upward and downward
indicate increases and decreases, respectively, upon UV-B treatment.
(b) Relative abundances of 13 and 10 significantly changed SalAs in leaves and roots, respectively, as calculated based on the obtained peak areas, and the resulting values in UV-B-exposed samples versus control were compared. Data represent means SDs of three independent biological replicates. Significant differences between UV-B-irradiated and control samples were determined using the Student t-test (*P < 0.05, **P < 0.01, ***P <0.001).
3. 蛋白质组学分析揭示了UV-B照射下与SalA生物合成直接相关的关键酶的变化
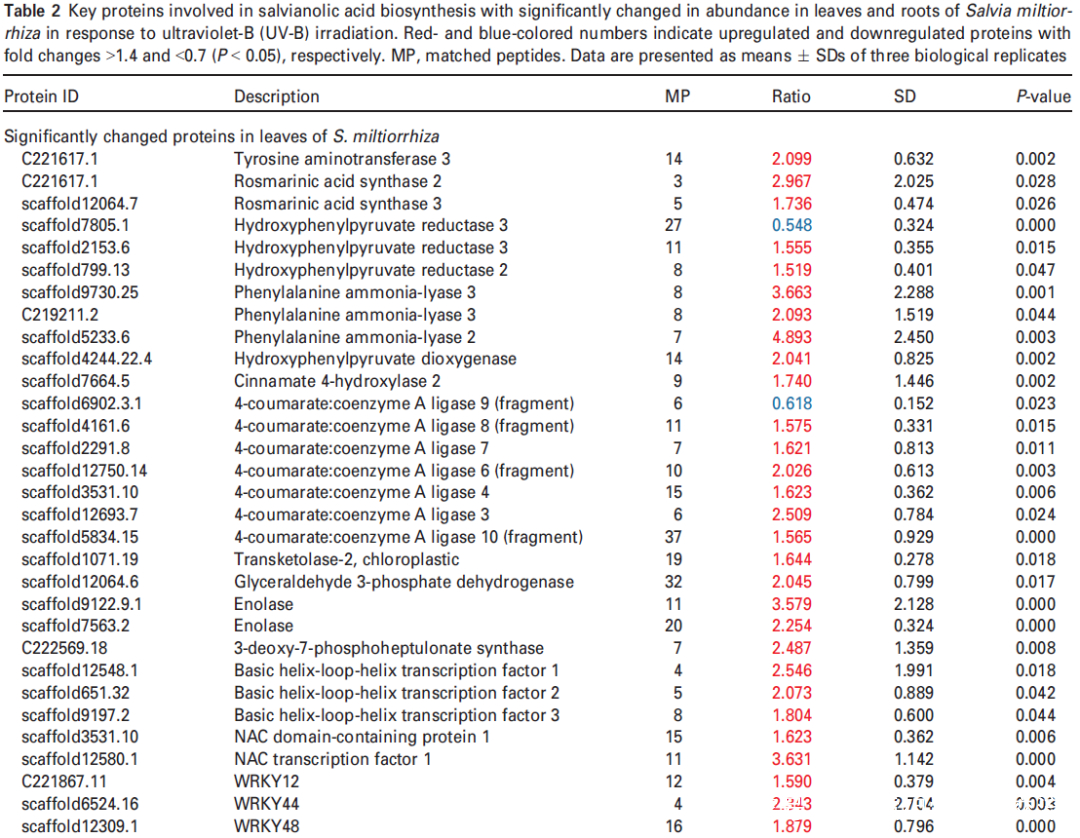
研究者进一步使用比较蛋白质组学分析探索了经UV-B照射诱导的SalA 生物合成机制。基于金开瑞公司的iTRAQ蛋白质组学方法对UV-B处理组和对照组样品的叶和根进行质谱检测分析。分析所有获得的肽段长度和unique肽的数量,以评估蛋白质组数据的质量,结果表明,鉴定出的肽段由8-38个氨基酸和2-20个unique肽段组成,使用置信度95%的过滤数据,在叶和根组织中总共鉴定出16830和21120个肽段,分别比对鉴定到的3303和3620个蛋白(图3(a))。根据Fold-Change大于1.4或小于0.7(P < 0.05)的筛选标准,鉴定到在UV-B照射下,丹参叶片中281个蛋白表达量显著上调,155个蛋白表达量显著下调。(图3(a);表S4),在丹参根中分别有102和157个蛋白表达量显著上调和下调(图3(a);表S5)。为支持蛋白质组学结果,经酶活性检测,在UV-B照射的丹参植物叶片中,经代谢组学检测分析得到的与SalA生物合成相关的关键酶苯丙氨酸氨裂合酶(PAL)、酪氨酸转氨酶 (TAT)等的活性也被确定显著增强(图3(c))。
进一步分析蛋白质组学数据,在UV-B处理的丹参叶子中,显示9种转录因子TF的蛋白表达水平升高,包括与拟南芥直系同源的bHLH1、bHLH2、bHLH3、NAC1、WRKY12、WRKY44、WRKY48、WRKY56和WRKY61,此外,发现与JA生物合成相关脂氧合酶(LOX) 和SA生物合成相关PAL蛋白表达水平显著升高在UV-B与对照相比(表2),与JA和SA的代谢积累一致(图S5和6;表S3)。在丹参根中,仅检测到两种TF的水平,即bHLH2和NAC1,在UV-B照射下显著增加,而UV-B照射后丹参根中JA生物合成相关酶的水平没有显著变化(表2)。
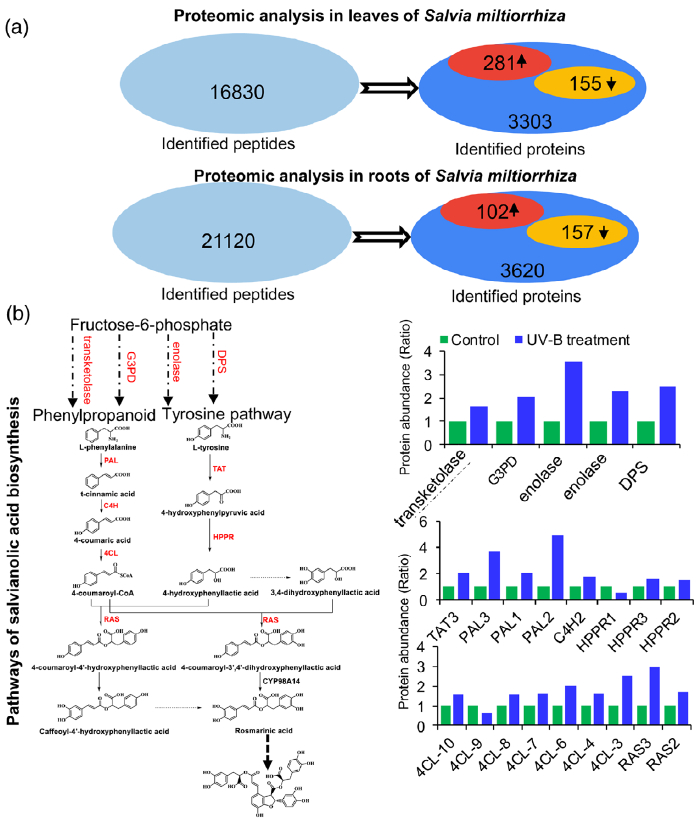
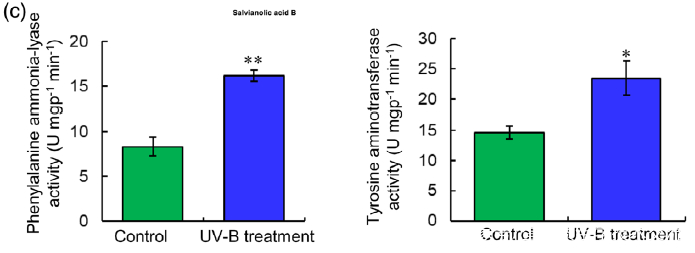
Figure 3. Proteomic analysis to identify key proteins involved in the biosynthesis of salvianolic acids (SalAs) under UV-B irradiation.
(a) Three-month-old Salvia miltiorrhiza plants were irradiated with UV-B for 4 h and compared with controls. The extracted proteins from leaves or
roots were subjected to a proteomic analysis. Differentially expressed proteins (DEPs) were identified by comparing protein levels in UV-B-irradiated versus control samples. Statistical analysis of identified peptides, proteins, and DEPs from leaves and roots, respectively, was performed. The arrows facing upward and downward indicate increases and decreases, respectively, due to UV-B exposure.
(b) (Left) Biosynthetic pathways of SalA from fructose-6-phosphate. SalA biosynthesis-related enzymes include transketolase, glyceraldehyde 3-
phosphate dehydrogenase (G3PD), enolase, 3- deoxy-7-phosphoheptulonate synthase (DPS), phenylalanine ammonia-lyase (PAL), tyrosine aminotransferase (TAT), cinnamate 4-hydroxylase (C4H), 4-coumarate:coenzyme A ligase (4CL), hydroxyphenylpyruvate reductase (HPPR), and rosmarinic acid synthase (RAS). Red color indicates proteins with increased abundance after UV-B irradiation. (Right) Protein abundance in control and treated samples.
(c) PAL and TAT enzymatic activities in control and UV-B-irradiated samples were measured. Data are presented as means SDs of three independent biological replicates. Significant differences between UV-B-irradiated and control samples were determined using the Student t-test (*P < 0.05, **P< 0.01).
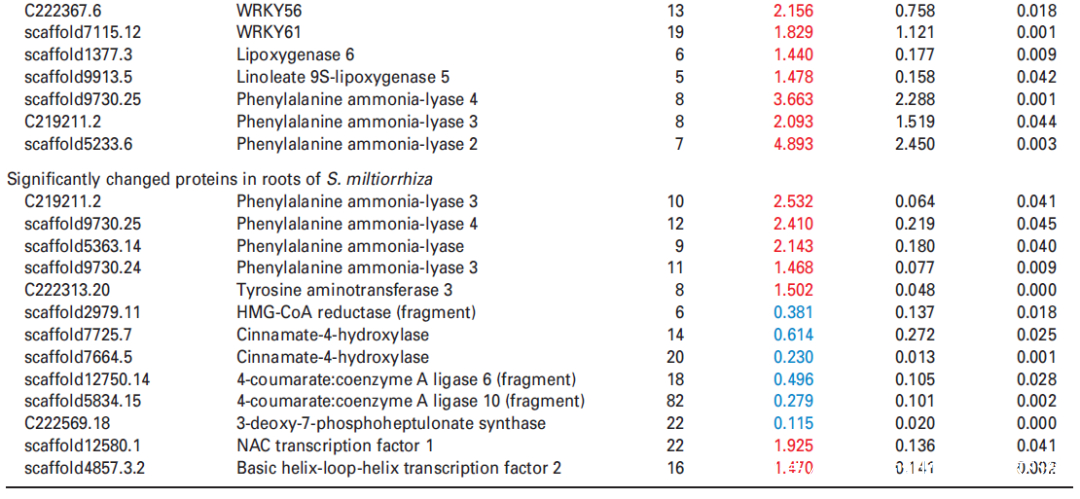
4. 蛋白组与代谢组关联分析
在本研究中,通过代谢组学分析发现,在UV-B照射下丹参叶片中有12种SalAs含量升高(图2(b);另一方面,蛋白质组学结果显示,与SalA生物合成、转录调控、蛋白质稳态(proteostasis)、光合作用、叶绿素合成和蔗糖代谢相关的蛋白质受UV-B照射影响较为显著(表2和S6)。进行蛋白组与代谢组关联分析,系统地评估了SalAs含量和UV-B应激蛋白水平变化之间的关系。如图4所示,根据与SalA含量的相关值,将显著变化的蛋白分为三个聚类。丹酚酸和迷迭香酸水平与定位在II簇的蛋白如SalA生物合成相关蛋白和TFs呈正相关;然而,它们与定位于簇I的蛋白呈负相关,包括光合作用相关蛋白(图4;表S7)。
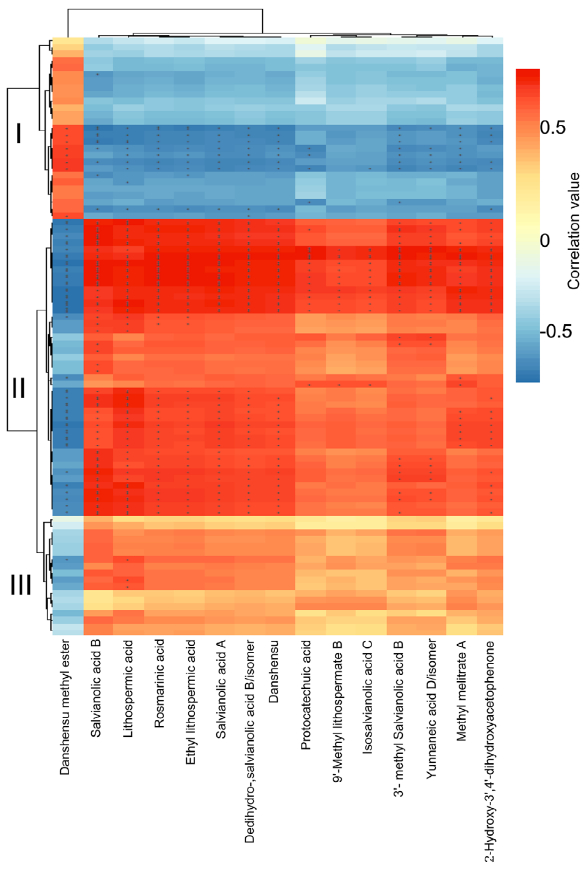
Figure 4. Correlation analysis between differentially expressed proteins and significantly changed salvianolic acids (SalAs). The use of the Pearson correlation coefficient enabled the establishment of the correlation between proteins that were significantly different in abundance and significantly changed SalAs. An asterisk indicates a significant correlation between differentially expressed proteins and differentially accumulated SalAs. Red and blue colors indicate positive and negative correlations, respectively. Detailed coefficient values are listed in Table S7.
5. NAC1在SalAs生物合成中的作用
研究者注意到在UV-B照射样本中,转录因子NAC1(或 SmNAC1)在丹参叶组织中表达量上调最为显著(图5(b),表明NAC1可能在SalA生物合成中起着关键调节作用。为了证明这一假设,使用含有带Flag标记的NAC1过表达质粒或带有绿色荧光蛋白(GFP)的NAC1-RNAi质粒的农杆菌Agrobacterium rhizogenes C58C1来合成过表达或沉默NAC1基因的转基因毛根NAC1-OE和NAC1-SI(图S8(a,b))。为了鉴定NAC1调控的基因,使用 NAC1-OE、NAC1-SI和对照组的毛根进行比较转录组分析。结果表明,SalA生物合成相关基因在NAC1-SI毛根中下调(表S8);PAL2的表达在NAC1-OE毛根中高度上调,而在NAC1-SI毛根中没有显著变化(表S8)。接下来,通过RT-qPCR评估了转基因毛状根中几种SalA生物合成相关基因的表达水平。正如预期的那样,与SalA生物合成相关的大部分基因和酶分别在NAC1-OE和NAC1-SI毛根中被显著诱导和抑制(图6(b))。聚类分析进一步证实了SalA生物合成相关基因的表达水平的变化和编码蛋白水平的变化显示出相似的趋势(图S9)。
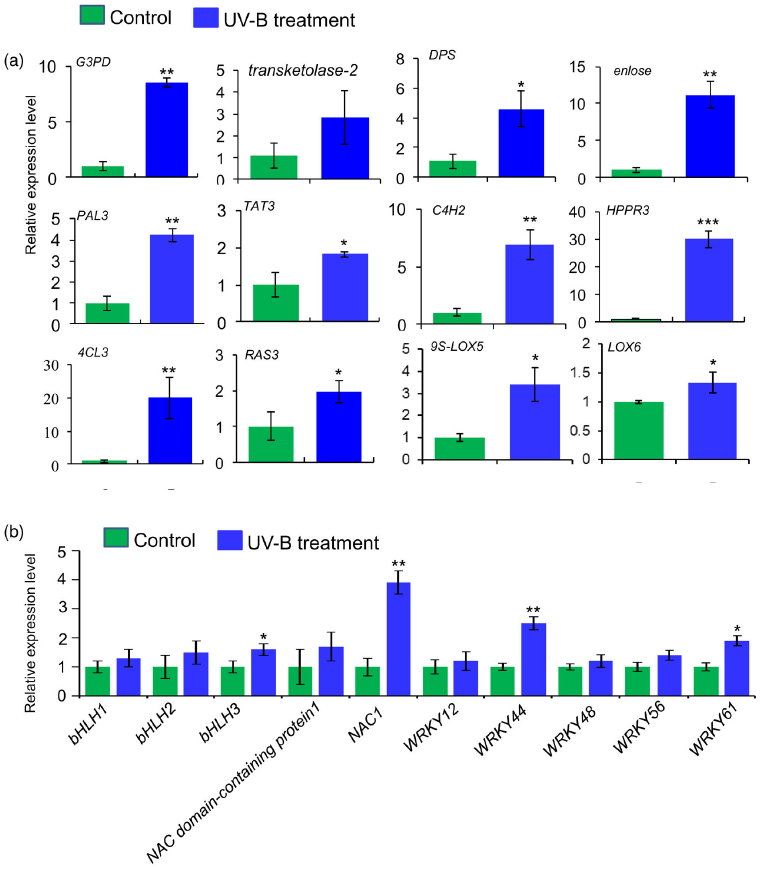
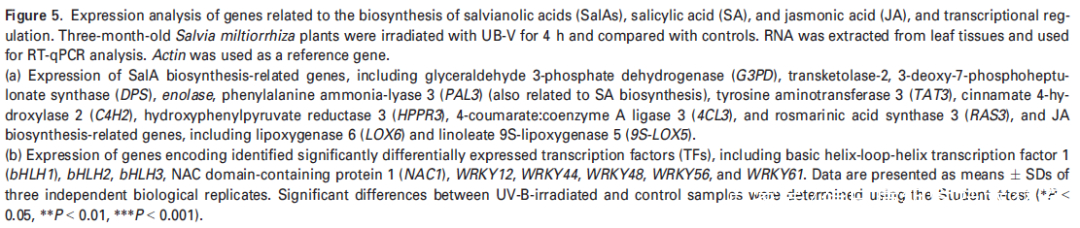
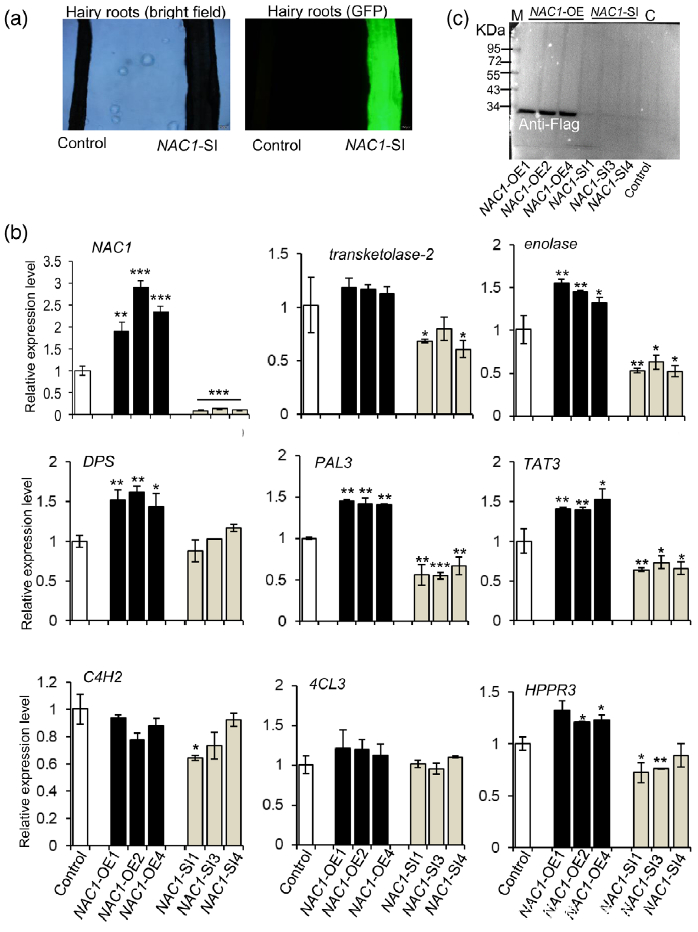
Figure 6. Overexpression or silencing of the NAC1 gene affects the expression of salvianolic acid (SalA) biosynthesis-related genes in hairy roots of Salvia miltiorrhiza.
(a) Green fluorescence protein (GFP) expression was observed by fluorescence microscopy to verify the presence of the NAC1-RNAi plasmid.
(b) Expression of genes related to SalA biosynthesis in NAC1-overexpressing (NAC1-OE) and NAC1-silencing (NAC1-SI) hairy roots. Data shown are means SDs of three independent biological replicates.
Significant differences between UV-B-irradiated and control samples were determined using the Student t-test (*P < 0.05, **P < 0.01, ***P <0.001).
(c) Increased NAC1 protein levels in NAC1-OE hairy roots as indicated by western blot analysis using anti-Flag. M, protein marker; C, control.
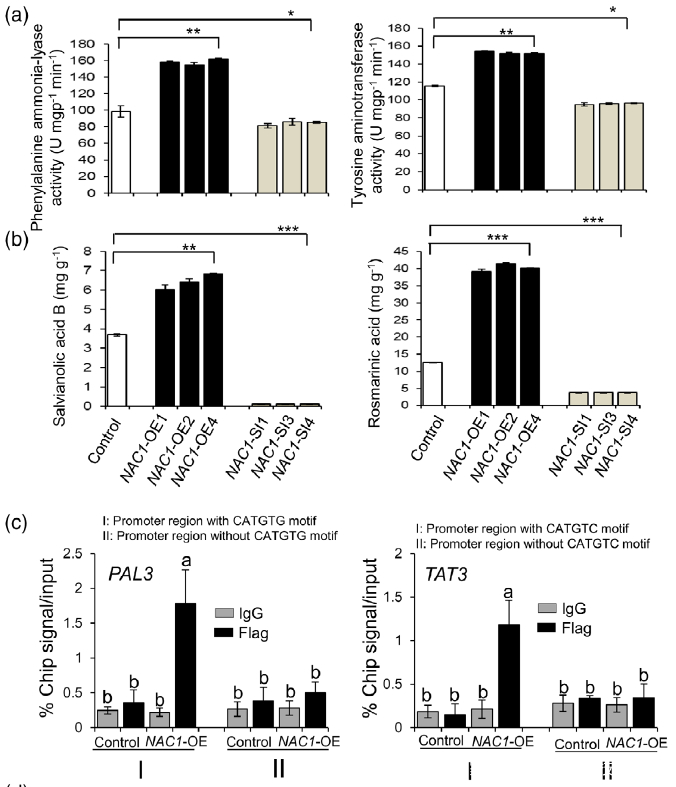
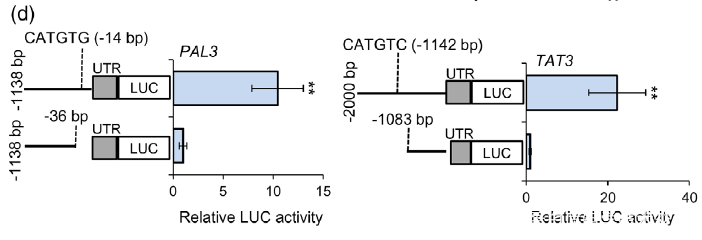
Figure 7. Effects of overexpression or silencing of NAC1 on the salvianolic acid B and rosmarinic acid contents.
(a) PAL and TAT enzymatic activities in NAC1-overexpressing (NAC1-OE) and NAC1-silencing (NAC1- SI) hairy roots.
(b) Rosmarinic acid and salvianolic acid B contents in NAC1-OE and NAC1-SI hairy roots. Data are expressed as means SDs of three independent biological replicates. Significant differences between UV-B-irradiated and control samples were determined using the Student t-test (*P < 0.05, **P< 0.01, ***P < 0.001).
(c) ChIP-qPCR assay using hairy roots of NAC1-OE and control lines. Immunoprecipitation (IP) was performed using an anti-Flag antibody or normal mouse immunoglobulin G (IgG) (negative control). PAL3 promoter regions with or without CATGTG cis-motif and TAT3 promoter regions with or without CATGTC cis-motif were comparatively amplified by RT-qPCR. The error bars indicate the SDs from three independent experiments. Different letters indicate statistical significance as analyzed using one-way ANOVA with Tukey’s multiple comparison (P < 0.05). TAT3, tyrosine aminotransferase 3; PAL3, phenylalanine ammonia-lyase 3.
(d) Dual-LUC assay showed the effect of NAC1 on the activation of PAL3 and TAT3 promoters. The promoters of PAL3 (with CATGTG) and TAT3 (with CATGTC) were individually fused to the luciferase (LUC) reporter gene. As negative control, the promoters of PAL3 and TAT3 genes without cis-motif were fused to the LUC gene. The promoter activities were determined by transient expression in tobacco. Relative LUC activity was normalized to the Renilla (REN) luciferase. Error bars indicate SD (n = 3). Significant differences were determined by the Student t-test (**P < 0.01).
四、总结
在这项研究中,研究者对暴露于4小时UV-B照射的丹参叶子和根进行了综合的代谢组学和蛋白质组学分析,代谢组学分析结果显示,经UV-B处理的丹参的叶和根中分别增加了12和9种SalAs。通过蛋白质组学分析获得的数据表明,暴露于UV-B辐射的丹参幼苗根中上调的SalA生物合成相关关键酶的数量(5种酶)远低于叶片中的(20种酶)(表2),并找到了SalA合成的关键转录调控因子NAC1。最后通过进一步的蛋白质组学分析、基因过表达和沉默、转基因实验等进一步验证了关键蛋白和基因的功能,并在分子水平上讨论UV-B诱导丹参SalA生物合成的机制。
